What if you could get even more out of your VITEK® 2 with the Advanced Expert System™ (AES)?
Here are three tips that you can use to boost efficiency:
- How to change an antibiotic interpretation in VITEK® 2 8.01 software for a given isolate
- How to display MIC distributions for each proposed phenotype and visually compare isolate MIC results
- How to use colored indicators to identify results that need attention
Need to print it to keep those tips and tricks close to your VITEK® 2?
How to change an antibiotic interpretation in VITEK® 2 8.01 software for a given isolate
Beginning with VITEK® 2 Systems 8.01 software you can now manage isolates on remote computers without the need to install software. Version 8.01 is the latest version of VITEK® 2 Systems software, and this version allows customers to manage isolates in either a web-based application mode (no software installation required) or the more traditional VITEK® 2 Systems software mode. Traditional VITEK® 2 Systems software can only be accessed by computers directly linked to the instrument itself, whereas the web-based VITEK® 2 Systems Web application is different. VITEK® 2 Systems Web application allows multiple users to setup ID and AST tests and access their results simultaneously from any networked PC with remote licenses applied. VITEK® 2 Systems Web application can also be accessed by computers directly linked to the instrument itself.
Changing an interpretation for an antibiotic when using VITEK® 2 Systems Web application is simple!
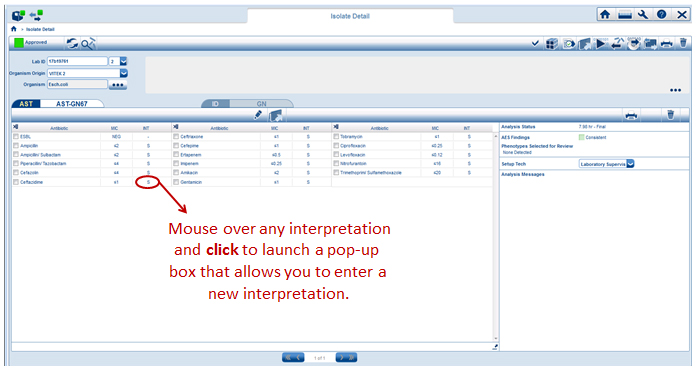
When in isolate detail view, as shown below, the system allows you to change MIC interpretations with a simple click of the mouse. Just mouse over the category interpretation you’d like to change and left-click the mouse as shown below.
The system will then create the drop-down box seen below that allows a different interpretation to be entered.
When running traditional VITEK® 2 Systems software (version 8.01 or previous versions) the process for changing an antibiotic interpretation is almost identical. In this case, when in isolate detail view, the system allows you to change an interpretation for an antibiotic with a simple Ctrl + click. Just mouse over the category interpretation you’d like to change and hold down the Ctrl key while left-clicking the MIC interpretation with your mouse. Now enter the new interpretation and you’re done! It’s that easy.
The following interpretations are available depending on your breakpoint parameter set configuration (e.g. CLSI, Global CLSI-based, EUCAST or Global European-based).
| S | Susceptible |
| I | Intermediate |
| R | Resistant |
|
IE |
Indicates that there is insufficient evidence that the species in question is a good target for therapy with the drug. A MIC with a comment, but without an accompanying S, I or R categorization may be reported. |
|
( – ) |
Indicates that susceptibility testing is not recommended as the species is a poor target for therapy with the drug. Isolates may be reported as R without prior testing.1 |
|
SDD |
Indicates that susceptibility is dependent on the dosing used. For isolates with MICs in the SDD range, it is necessary to use higher doses, more frequent doses, or both. For more information, refer to the CLSI M100 document. |
1) The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 7.1, 2017, http://www.eucast.org/fileadmin/src/media/PDFs/
EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf
How to display MIC distributions for each proposed phenotype and visually compare isolate MIC results
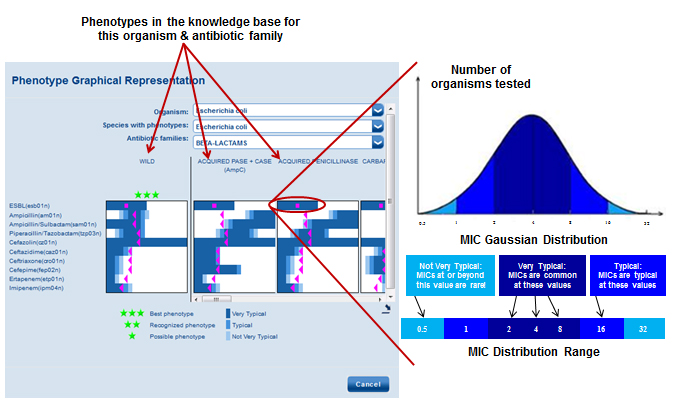
Click the cube icon (![]() ) to access the AES Detail Screen. Here you will find a list of phenotypes in the knowledge base for the organism in question. Phenotypes are viewed by antibiotic family and you can toggle between antibiotic families using the convenient drop-down box. For each phenotype, a histogram is shown illustrating the expected MIC distributions for each antimicrobial tested (See Figure 1). Proposed matches are indicated by green stars. Pink squares and triangles representing the isolate’s MICs for a given antimicrobial are plotted for each phenotype to show how MIC results for the isolate tested compare to the expected MIC distributions of each phenotype (See Figure 2).
) to access the AES Detail Screen. Here you will find a list of phenotypes in the knowledge base for the organism in question. Phenotypes are viewed by antibiotic family and you can toggle between antibiotic families using the convenient drop-down box. For each phenotype, a histogram is shown illustrating the expected MIC distributions for each antimicrobial tested (See Figure 1). Proposed matches are indicated by green stars. Pink squares and triangles representing the isolate’s MICs for a given antimicrobial are plotted for each phenotype to show how MIC results for the isolate tested compare to the expected MIC distributions of each phenotype (See Figure 2).
Figure 1 – Click to enlarge
Figure 2 – Click to enlarge
How to use colored indicators to identify results that need attention
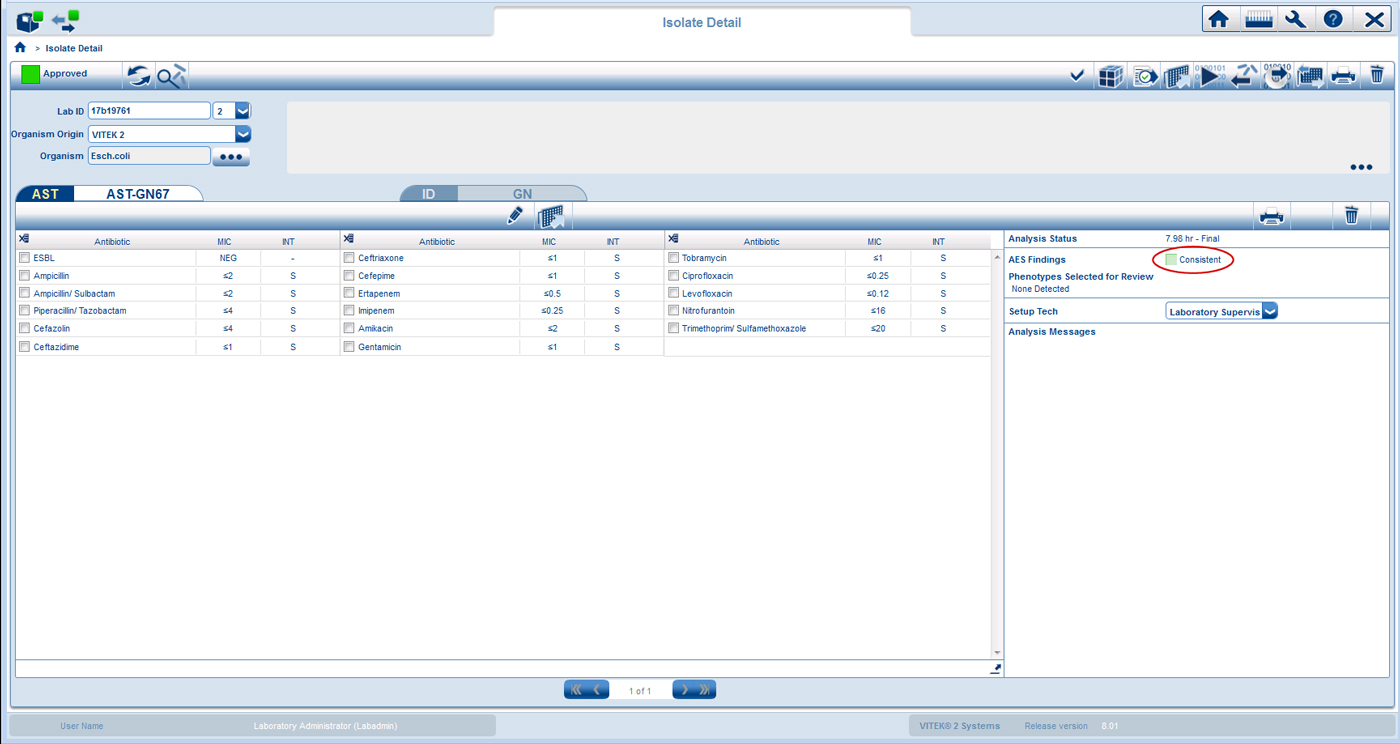
The VITEK® 2 color codes every isolate to indicate the quality of the match to a known phenotype in the database. These color codes are referred to as “AES Findings”, and they are found in isolate detail view as shown below.
![]() All MICs of the identified isolate match the expected MIC distribution for a defined phenotype. The AES knowledge base is very robust covering 141 organisms, and 4,000 phenotypes comprised of 55,000 MIC distributions2.
All MICs of the identified isolate match the expected MIC distribution for a defined phenotype. The AES knowledge base is very robust covering 141 organisms, and 4,000 phenotypes comprised of 55,000 MIC distributions2.
![]() One MIC value does not fit the expected pattern for a known phenotype for the tested organism. Consistent result with biologic correction.
One MIC value does not fit the expected pattern for a known phenotype for the tested organism. Consistent result with biologic correction.
![]() The isolate could not be matched to a known phenotype because the MIC values of more than 1 antimicrobial does not match a defined MIC distribution in the database.
The isolate could not be matched to a known phenotype because the MIC values of more than 1 antimicrobial does not match a defined MIC distribution in the database.
![]() AST results are available, because the organism is claimed by the AST knowledge base, but AES expert analysis is not performed since the species identified is not claimed in the VITEK® 2 AES database. No data in the Advanced Expert System™ knowledge base.
AST results are available, because the organism is claimed by the AST knowledge base, but AES expert analysis is not performed since the species identified is not claimed in the VITEK® 2 AES database. No data in the Advanced Expert System™ knowledge base.
![]() No AST results or AES expert analysis, because none of the antibiotics have defined breakpoints.
No AST results or AES expert analysis, because none of the antibiotics have defined breakpoints.
Need to print it to keep those tips and tricks close to your VITEK® 2?
References:
-
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 7.1, 2017, http://www.eucast.org/fileadmin/src/media/PDFs/ EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf
-
VITEK® 2 8.01 software